The last 20 years have brought a revolution in targeted therapies for cancer. Small-molecule inhibitors and monoclonal antibodies that target a specific aberrant protein in tumors have provided cancer patients with treatments that are associated with fewer side effects and longer survival than conventional chemotherapy. This has been, in large part, the result of intensive research into the role of oncogenes in cancer development. Oncogenes are normal cellular genes that have become mutated in such a way that they aberrantly promote the uncontrolled cell growth seen in cancer. They are often proteins involved in growth control or activation of cellular signaling; inhibiting these mutated proteins has proven to be effective in stopping the growth of many cancers. Research by a team from the Brown Cancer Center at the University of Louisville in Kentucky using the U.S. Department of Energy’s Advanced Photon Source (APS) and published in the journal Nucleic Acids Research promises to extend these treatment possibilities to control these oncogenes at the gene expression level. The work, based on the discovery that DNA in the promoter region of many genes forms higher order structures that could provide unique druggable targets for intervention, extends structural knowledge of the promoter regions of three important oncogenes.
The basis for the work comes from previous studies which showed that DNA and RNA sequences containing four stretches of three guanine nucleotides separated by 1-7 additional nucleotides could form stacked secondary structures called G4 quadruplexes (G4s). These sequences are frequently observed in the promoter region of genes at locations known to interact with transcription factor proteins that regulate gene expression. In addition, experiments have shown that these G4 regions act as transcription factor “hubs” and that small molecule inhibitors can disrupt these interactions, making them a potential site for drug intervention. However, due to the limitations of conventional structural biology methods and the heterogeneity and complexity of these structures, most G4 structure studies have relied on short, truncated G4 sequences (< 33 nucleotides) that have exhibited few unique features and may not represent the true biological context. These researchers hypothesized that longer sequences should form higher order structures with sufficient complexity and specificity to provide unique drug targets.
In order to address the limitations of previous studies, the team developed an iterative integrated structural biology platform that enabled them to combine structural biology and biophysical methods such as circular dichroism (CD), analytical ultracentrifugation sedimentation velocity (AUC-SV), and small-angle x-ray scattering (SAXS) together with molecular modeling to study higher order structures of long (34-70 nucleotides) G4 sequences in a homogeneous, physiologically relevant solution.
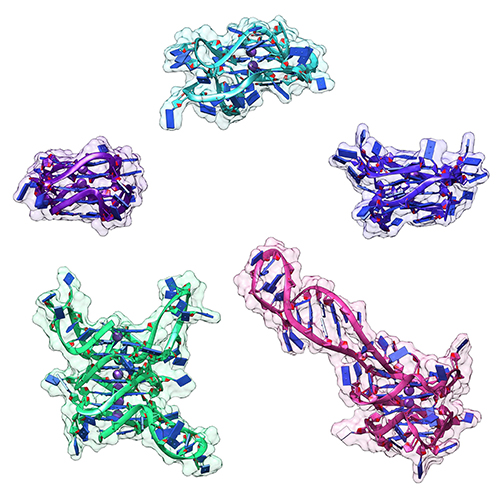
Their results provide a number of important advances that should aid drug discovery for cancers that are driven by aberrations in regulation of three major oncogenes, c-Myc, c-Kit, and k-Ras. First, CD analyses revealed that the team could enumerate the number of G4 stacks in their long oncogene promoter-derived sequences. These structural predictions were reinforced y AUC-SV analyses which also showed that the long promoter sequences form true compact secondary structures and not just random coils. SAXS analysis, which was performed at the Biophysics Collaborative Access Team (Bio-CAT) 18-ID beamline at the APS, provided additional structural information consistent with the other analyses and showed the G4s to be folded, compact, globular structures resembling proteins more than DNA. The APS is an Office of Science user facility at Argonne National Laboratory.
Molecular modeling of the structures was constrained by the data from the CD analyses and then fit into refined SAXS envelopes, providing atomistic models (Fig. 1) needed for drug discovery activities. In fact, based on modeling against other known drugs and their druggable targets, these higher order structures for the promoter regions of the three oncogenes provide enough structural complexity and uniqueness to be good-to-excellent druggable targets.
The next step for the team will be to use their own drug discovery platform to identify possible drug candidates for these oncogenic targets, an important advancement that will have significant importance in the clinic.
See: Robert C. Monsen, Lynn W. DeLeeuw, William L. Dean, Robert D. Gray, Srinivas Chakravarthy, Jesse B. Hopkins, Jonathan B. Chaires and John O. Trent, “Long promoter sequences form higher-order G-quadruplexes: an integrative structural biology study of c-Myc, k-Ras and c-Kit promoter sequences,” Nucleic Acids Res. 50 (7), 4127 (2022). DOI: 10.1093/nar/gkac182